Frontiers in Plant Science, 13: 992543, 2022
Abstract
Heterotrimeric Nuclear Factor Y (NF-Y) transcription factors are key regulators of the symbiotic program that controls rhizobial infection and nodule organogenesis. Using a yeast two-hybrid screening, we identified a putative protein kinase of Phaseolus vulgaris that interacts with the C subunit of the NF-Y complex. Physical interaction between NF-YC1 Interacting Protein Kinase (NIPK) and NF-YC1 occurs in the cytoplasm and the plasma membrane. Only one of the three canonical amino acids predicted to be required for catalytic activity is conserved in NIPK and its putative homologs from lycophytes to angiosperms, indicating that NIPK is an evolutionary conserved pseudokinase. Post-transcriptional silencing on NIPK affected infection and nodule organogenesis, suggesting NIPK is a positive regulator of the NF-Y transcriptional complex. In addition, NIPK is required for activation of cell cycle genes and early symbiotic genes in response to rhizobia, including NF-YA1 and NF-YC1. However, strain preference in co-inoculation experiments was not affected by NIPK silencing, suggesting that some functions of the NF-Y complex are independent of NIPK. Our work adds a new component associated with the NF-Y transcriptional regulators in the context of nitrogen-fixing symbiosis.
Keywords: protein kinase, nitrogen fixation, signaling, symbiosis, transcription factor
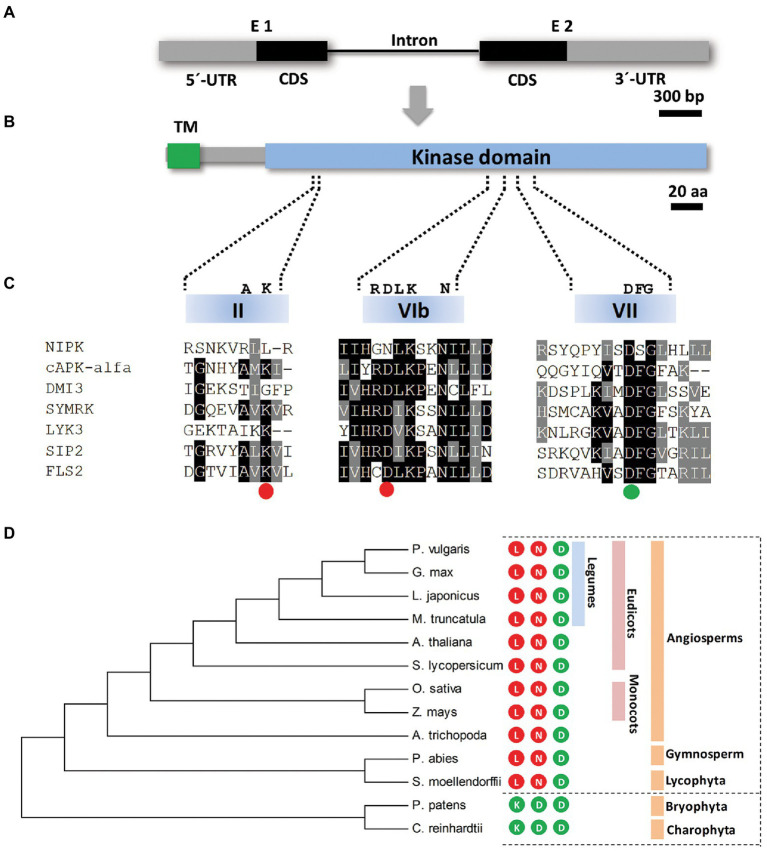
NIPK encodes a protein pseudokinase. (A,B) Schematic representation of the NIPK gene (A) and the encoded protein (B). Gray boxes correspond to 5′ and 3′ untranslated regions (UTR) and black boxes correspond to coding sequence regions. The black line indicates the only intron present in the NIPK gene. The putative transmembrane (TM) and the kinase domains of the protein are shown in green and light blue, respectively. (C) A multiple sequence alignment of motives VAIK (II subdomain), HRD (VIb subdomain), and DFG (VII subdomain) of the kinase domain of NIPK and the functional kinases DMI3 (Gleason et al., 2006), SYMRK (Yoshida and Parniske, 2005), LIK3 (Jayaraman et al., 2017), SIP2 (Chen et al., 2012), FLS2 (Lu et al., 2010), and cPKA-alfa. The colored circles indicate whether each of the three amino acids (K, D, and D) required for the phosphotransfer reaction are conserved (green) or not (red) in NIPK. (D) Phylogenetic tree generated with the amino acidic sequences of P. vulgaris NIPK and its putative orthologs from Medicago truncatula, Arabidopsis thaliana, Lotus japonicus, Glycine max, Amborella trichopoda, Solanum lycopersicum, Zea mays, Oryza sativa, Selaginella moellendorffii, Picea abies, Physcomitrella patens, and Chlamydomonas reinhardtii. The phylogenetic tree was generated using MEGA7. Numbers represent bootstrap values obtained from 1,000 trials. The colored circles indicate whether each of the three amino acids required for the phosphotransfer reaction (K, D, and D) are present (green) or not (red) in the amino acid sequence of NIPK orthologs in each species.